New data highlight on large-scale Gene Regulatory Network (GRN) simulation with perturbation-aware single-cell data
September 18, 2025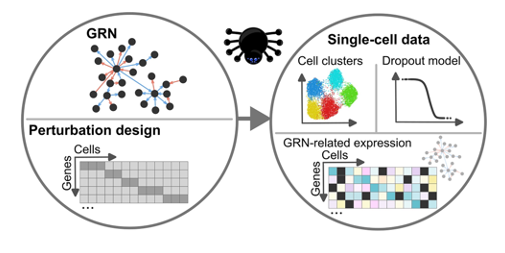
The latest data highlight, entitled ‘Large-scale Gene Regulatory Network (GRN) simulation and benchmarking using perturbation-aware single-cell data’, is based on recent research by Garbulowski et al. (2024), published in NAR Genomics and Bioinformatics. This work presents GeneSPIDER2, an open-source MATLAB toolbox designed to simulate large, biologically realistic gene regulatory networks (GRNs) and to benchmark network inference methods using synthetic single-cell data with perturbations.
GeneSPIDER2 introduces a number of advances over its predecessor, including support for generating scale-free and modular GRNs with thousands of genes, and for producing synthetic single-cell RNA-seq data that incorporates CRISPR-like knockdown perturbations. The tool allows researchers to control signal-to-noise ratios, dropout effects, and clustering, mimicking the characteristics of experimental data from protocols such as Perturb-seq and CROP-seq. The toolbox also integrates wrappers for inference algorithms like GENIE3, TIGRESS, and PLSNET—making it a flexible platform for method development and evaluation. Benchmarking results demonstrate that synthetic data generated by GeneSPIDER2 replicates key statistical properties and perturbation effects seen in real datasets from K562 and Calu-3 cell lines. These capabilities position the tool as a valuable resource for the systems biology and network inference communities.
To find out more, read the full data highlight: ‘Large-scale Gene Regulatory Network (GRN) simulation and benchmarking using perturbation-aware single-cell data’.
If you would like to showcase your research involving open data, software, or tools, please get in touch with us.