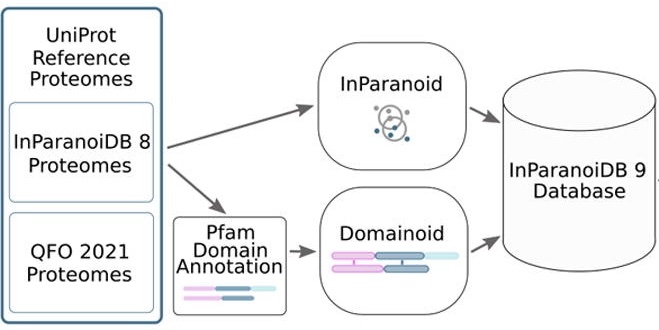
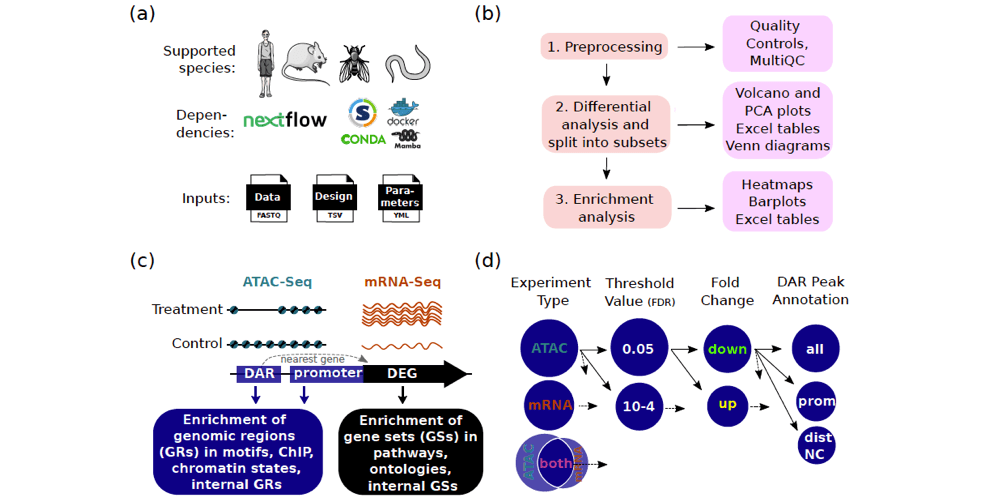
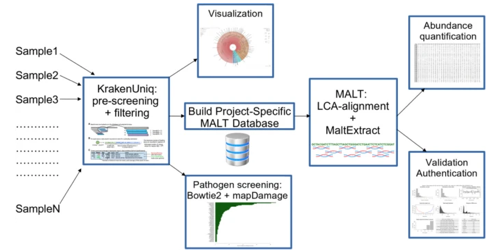
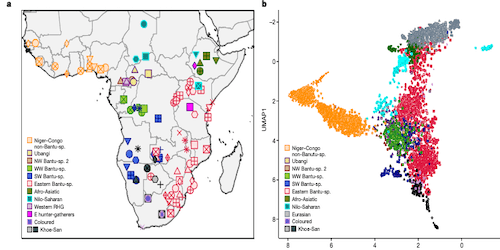
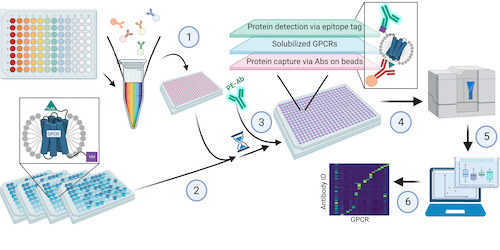
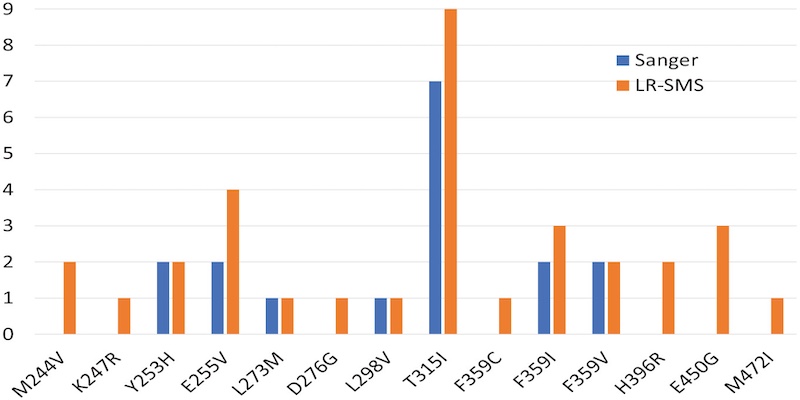
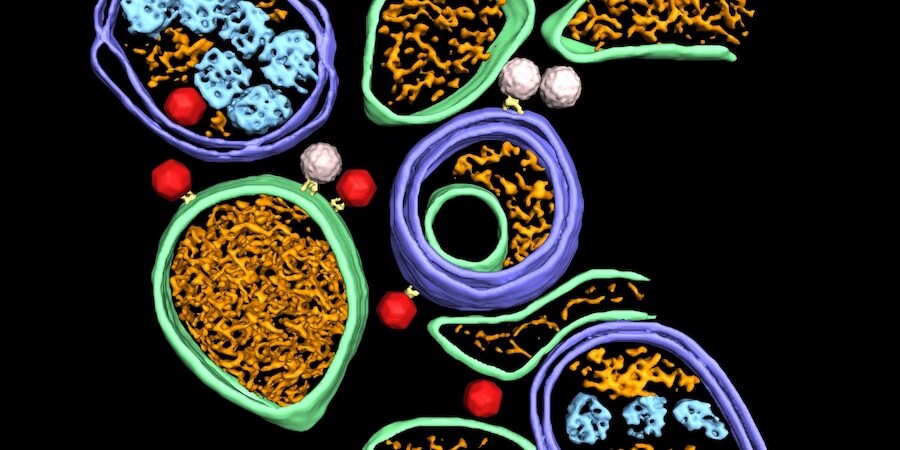
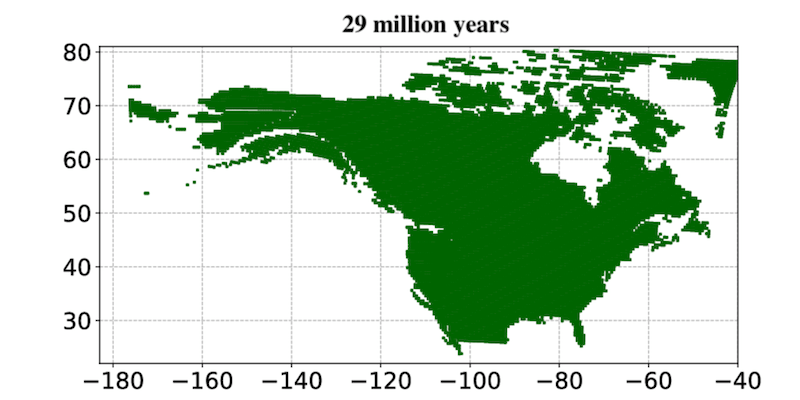
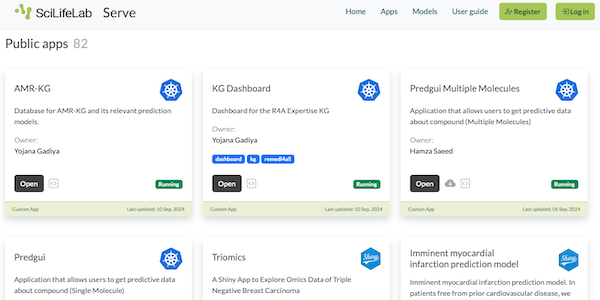
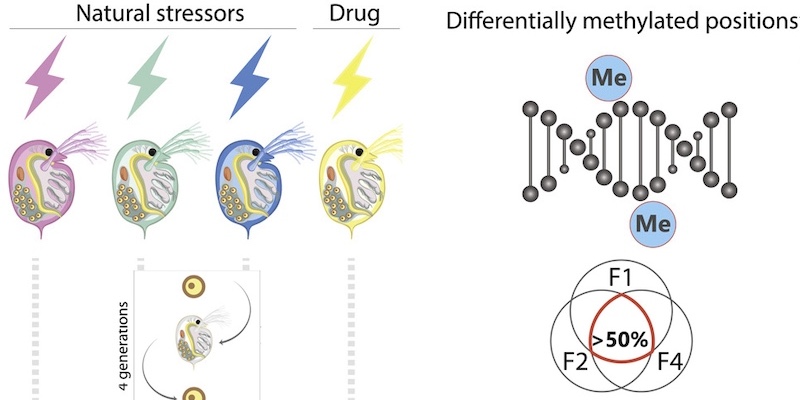
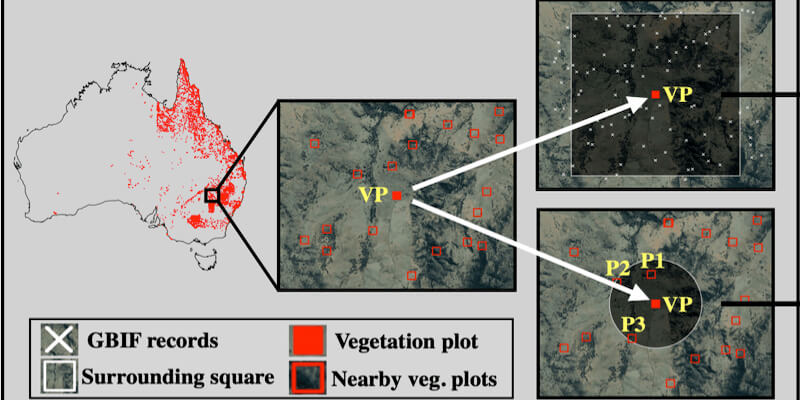
This page displays news relevant to data-driven life science in Sweden, e.g. open calls and large research efforts. It will also show news related to the SciLifeLab Data Platform. For example, we will announce updates on the tools, services, and databases on the SciLifeLab Data Platform.
May 20, 2025
April 15, 2024
January 19, 2024
January 11, 2024
November 15, 2023
September 29, 2023
September 7, 2023
August 8, 2023
July 4, 2023
April 28, 2023
April 18, 2023
March 24, 2023
March 3, 2023
December 22, 2022
December 19, 2022
December 9, 2022
October 21, 2022
June 2, 2022