Large-scale Gene Regulatory Network (GRN) simulation and benchmarking using perturbation-aware single-cell data
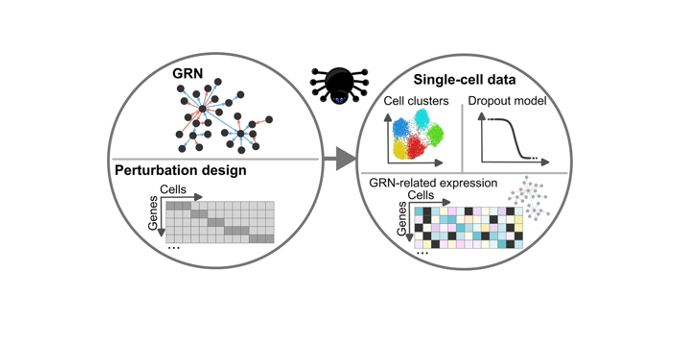
Gene regulatory networks (GRNs) are crucial for understanding gene expression control in biological systems. The emergence of single-cell transcriptomics has transformed GRN inference by offering detailed insights into cellular regulation at the individual cell level. However, computational challenges persist in evaluating the accuracy of GRN inference tools, particularly when it comes to simulating realistic data that include genetic perturbations. This is a growing focus in systems biology due to its applications in cancer research and immunology, including studies on SARS-CoV-2 response mechanisms.
Simulated datasets are commonly used for benchmarking GRN inference methods but, until now, most single-cell data simulators could not incorporate the effects of genetic perturbations or scale efficiently to large, biologically realistic networks. GeneSPIDER2 addresses these limitations by offering a powerful MATLAB-based framework tailored for large-scale, perturbed single-cell simulations.
In a recent publication in NAR Genomics and Bioinformatics, researchers from Stockholm University, Uppsala University, Karolinska Institutet, University of Wisconsin-Madison, and National Cheng Kung University introduced GeneSPIDER2. GeneSPIDER2 is a substantial update to their previously released GeneSPIDER toolbox for GRN benchmarking. The goal of the research was to develop an advanced and scalable simulation tool that can model realistic single-cell gene expression data, including perturbations, and benchmark the performance of GRN inference methods using this synthetic data.
GeneSPIDER2 introduces several significant enhancements compared previous GRN interference tools:
- Scalability: Ability to simulate GRNs with up to 20,000 genes using a novel modular stitching algorithm based on the Barabási–Albert model.
- Perturbation-aware simulation: First simulator to incorporate knockdown-based genetic perturbations into single-cell data.
- Realism: Synthetic data accurately mimics real CRISPRi Perturb-seq datasets from K562, Calu-3, HCC38, and HCC1143 cell lines in terms of dropout rates, expression distributions, and cluster structures.
- Flexibility: Includes tunable parameters for signal-to-noise ratio (SNR), cluster variance, and dropout modeling.
- Benchmarking-ready: Bundles a collection of inference methods (e.g. GENIE3, ARACNE) and allows comprehensive evaluation of GRN reconstruction accuracy.
GeneSPIDER2 was developed to fill a critical gap in benchmarking GRN inference using realistic and scalable simulations. By enabling perturbation modeling in single-cell data, the authors aim to bring researchers one step closer to understanding complex regulatory mechanisms in health and disease. GeneSPIDER2 holds potential to become a standard tool for data-driven investigations across disciplines.
GeneSPIDER2 delivers a highly relevant contribution to the field of transcriptomics and computational biology. Its unique capacity to simulate perturbation-aware single-cell datasets aligns well with the objectives of data platforms supporting systems medicine, pathogen research, and pandemic preparedness. By offering a reliable benchmarking environment, GeneSPIDER2 supports the development and evaluation of inference tools that can decipher regulatory processes underlying infection responses and disease states. As perturbation-based datasets grow in volume and importance, tools like GeneSPIDER2 will be central to extracting meaningful biological insights and informing precision health strategies.
Article
Garbulowski, M., Hillerton, T., Morgan, D., Seçilmiş, D., Sonnhammer, L., Tjärnberg, A., Nordling, T.E.M, & Sonnhammer, E. L. (2024). GeneSPIDER2: large scale GRN simulation and benchmarking with perturbed single-cell data. NAR Genomics and Bioinformatics, 6(3), lqae121.
Funders
Science for Life Laboratory’s SFO program (M.G.); National Science and Technology Foundation of Taiwan (NSTC 112-2314-B-006-079 to T.E.M.N.). Funding for open access charge: Stockholm University.
Data Availability
- GeneSPIDER2 runs under the MATLAB computing platform and is available under GPLv3 license at Zenodo.
- Software source code and postprocessed fold-change gene expression data with perturbation design matrices are publicly available at Bitbucket.
- Raw single-cell RNA-seq data were uploaded on Figshare and GEO (GSE208240 and GSE241115).