Large scale immunofluorescence to explore the host cell response to SARS-CoV-2 infection
Over the last two years, the COVID-19 pandemic has significantly affected people’s lives worldwide. Apart from vaccination to prevent COVID-19, there is a need to identify different treatment options for people already being infected and suffering from it. Further, general strategies for how to identify and develop effective drugs for any cause of possible future pandemics are needed.
The process of drug development, from identifying the compound to an approved drug is both long and costly. Therefore, drug repurposing – using already approved drugs for treatments of another condition, is a way to shorten this time frame. However, thousands of drugs are available, and it would not be practically possible to test them all for every new possible purpose. To make drug repurposing more effective and less resource demanding, a targeted approach could help determining which drugs are potentially usable.
One strategy that has been suggested to ‘narrow down’ the number of drugs for drug repurposing is based on identifying the host cell proteins that change location or abundance upon infection in situ using immunofluorescence. These proteins are considered to have a role in the viral replication life cycle and are thereby putative targets to treat infection. Once such proteins are identified, drugs that target those proteins could be applied in drug screening and evaluated for their effect on antiviral activity. This strategy was recently explored by researchers from KTH/SciLifeLab and Karolinska Institute/Scilifelab. In the preprint recently released by the researchers (first author: Jayasankar Mohanakrishnan Kaimal and Marianna Tampere, corresponding authors: Päivi Östling and Charlotte Stadler), they demonstrate that this strategy is feasible.
The researchers developed an assay for SARS-CoV-2 infection of Vero-E6 cells. Then, the subcellular localisation and expression of ≈ 600 host proteins were evaluated in mixed populations of infected and non-infected cells using immunofluorescence and antibodies from the Human Protein Atlas (HPA) project.
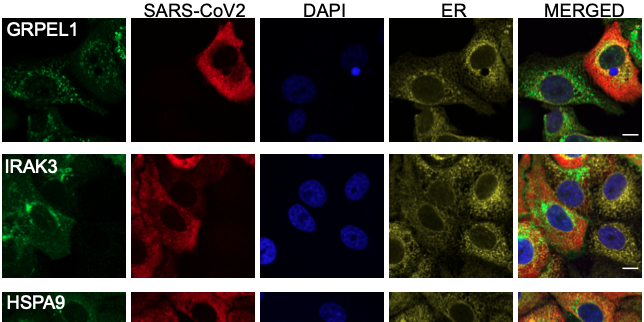
Among the 602 proteins, 97 were found to have an altered profile in infected cells. Of these, 45 were shown to have elevated abundance whilst 10 showed reduced abundance. Further, 20 proteins changed their localization within the cell when infected, and 22 proteins exhibited changed expression and localisation.
The host cell proteins identified in the assay were then considered as targets for a drug repurposing screen, using compounds from the SPECS repurposing library (Broad Institute). The team thus further optimized their host response screen to be compatible with drug repurposing in SARS-CoV-2 infected cells. In total, 116 drugs towards 21 of the identified host targets were available from the SPECS library and included in the repurposing screen. From the screen three compounds - elesclomol, crizotinib and rimcazole – were identified with significant antiviral activity and will be subjects for further validation studies. In summary, the study demonstrates that immunofluorescence used to identify potential host targets, is an effective approach for more targeted drug repurposing. This could have significant impact in the future by enabling researchers to ‘narrow down’ the number of existing drugs that are likely to be useful for treatment, or to identify novel targets for drug discovery.
Immunofluorescence images
The team has openly shared images ≈75,000 confocal images of SARS-CoV2 infected and non-infected Vero E6 cell populations under DOI:10.17044/scilifelab.14315777:
Cells had been infected with the virus for 1 h and fixed 24 hour post infection in 96 well plates. Using immunofluorescence (4 colours) cells were stained for the SARS-CoV2 virus (red), the endoplasmic reticulum (yellow) and nucleus (blue) – same for all images in the data set and used as reference markers. On top of this, 700 different target proteins were stained – one per well (green). Images were acquired with an Opera Phoenix microscope at 63X, using 9 FoV and 3 z-planes per protein target.
Explore the images
Below, you can explore the immunofluorescence images using the Vizarr image viewer which allows instant visulization of large image dataset. It is powered by ImJoy.
Select a plate to view using the dropdown below. You can see the HPA ID of the antibody used and protein or gene name on the top left corner of the window. Scroll images using the slider just below or the three dots to the right of antibody ID to type a particular image number. Further down, you can adjust viewing options including changing contrast of each image channel.
Preprint
Kaimal, J. M., Tampere, M., Le, T. H., Axelsson, U., Xu, H., Axelsson, H., Bäckström, A., Marabita, F., Moussaud-Lamodière, E., Njenda, D., Sepulveda, C. O., Oyuang, W., Seashore-Ludlow, B., Vernersson, C., Mirazimi, A., Lundberg, E., Östling, P., Stadler. C. (2022). Subcellular mapping of the protein landscape of SARS-CoV-2 infected cells for target-centric drug repurposing. bioRxiv. 10.1101/2022.03.29.482838.