Research into the immunogenicity of GPCR epitopes to enhance future drug development
Proteins in the GPCR superfamily (heterotrimeric guanine-nucleotide-binding regulatory protein (G protein)–coupled receptors) are important in normal cellular physiology and intercellular communication. Disruptions of GPCR signalling are connected to several diseases, such as retinitis pigmentosa (rhodopsin mutations), nephrogenic diabetes insipidus (vasopressin receptor mutations), and obesity (melanocortin receptor mutations).
GPCRs are the targets of about a third of all approved therapeutic drugs, and are therefore important in developing and repurposing therapeutic drugs. Recently, biological drugs that modulate GPCR function based on the binding of anti-GPCR antibodies were introduced, e.g. erenumab, a Calcitonin gene-related peptide (CGRP) -R antagonist for treating migraines (Sweden, Aimovig (c)). However, challenges remain that must be addressed. For example, validating the selectivity of anti-GPCR antibodies across a broader range of GPCRs is difficult because there is a high sequence similarity between GPCRs in the same subfamily. In addition, anti-GPCR antibodies are critical reagents in many bioanalytical methods used to study GPCR biology and pharmacology. Therefore, high-throughput methods are needed to study GPCRs and specific antibodies, in in order to support future pharmaceutical drug development.
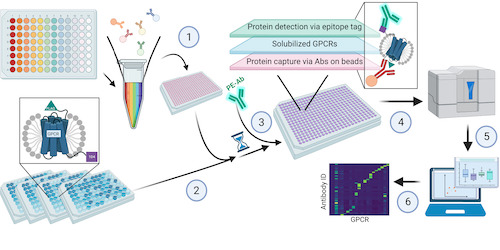
In a 2023 article in Science Advances, researchers from KTH Royal Institute of Technology, The Rockefeller University in New York, and Stockholm University (First author: Leo Dahl, Ilana B. Kotliar, Annika Bendes, Corresponding author: Jochen M. Schwenk, Thomas P. Sakmar) addressed the challenge of developing a high-throughput method to study GPCRs. Spearheaded by the work of three Ph.D. students (Dahl, Kotliar, and Bendes), the team established a framework based on multiplex immunoassays to evaluate the selectivity of anti-GPCR antibodies. A customised library of 215 expressed and solubilised GPCRs from all subfamilies was developed using dual epitope–tagged GPCRs in Expi293F cells (a special kidney HEK 293T cell line). Built on a previous proof-of-concept study, the team profiled over 400 anti-GPCR antibodies from the Human Protein Atlas.
The researchers used a suspension bead array (SBA) assay and a set of solubilised GPCR constructs with genetically encoded epitope tags. Each SBA contained antibodies that were covalently coupled to a distinct population of colour-coded magnetic beads. Antibodies for a specific GPCR subfamily and the epitope tags were then pooled into subfamily-specific SBAs. These multiplexed SBA assays exposed all antibodies to one GPCR at a time. This meant that every GPCR served as an on-target for some antibodies and as a probable off-target for all other antibodies. The researchers used the engineered epitope tags of the immunocaptured proteins to determine relative quantities of the GPCRs in solution. All data was added into a web-based interface that allowed the researchers to browse the selectivity of each antibody and GPCR. The team then used structural prediction models to interpret the on- or off-target binding events.
The results showed that almost two thirds of the antibodies tested were selective for their intended target, one in ten bound off-target, and one third did not bind to any GPCR. In addition, the analysis indicated that the antigens of on-target antibodies, on average, were longer, more disordered, and less likely to be buried in the interior of the GPCR protein compared to other antibodies. The researchers shared this data. They also presented a web-based R Shiny app that allows others to browse the assay results (from both antibody- and GPCR- centric perspectives), and also includes information about GPCR expression and enrichment of GPCR per antibody.
In summary, Dahl, Kotliar, and Bendes et al. provide important insights into GPCR immunogenicity. The researchers openly shared their data and developed an R Shiny app to enable the community to browse and analyse the data. In addition, the researchers proposed that their workflow could also be used to test other antibodies, such as nanobodies or scaffold proteins, if these can be immobilised to the beads. These results may, in the future, aid the development of more specific therapeutic drugs and agents.
Data
-
Access the openly shared data.
-
Access the web-based R app.
-
The code used for analysis and visualisation of the data is available in two Zenodo repositories: Multiplexed selectivity screening of anti-GPCR antibodies and GPCR structure analysis.
Article
Dahl, L., Kotliar, I.B., Bendes, A., Dodig-Crnković, T., Fromm, S., Elofsson, A., Uhlén, M., Sakmar, T.P., Schwenk, J.M. (2023). Multiplexed selectivity screening of anti-GPCR antibodies. In: Sciene Advances (Vol. 9, Issue 18, eadf9297).
Funding
This study was funded by the Knut and Alice Wallenberg Foundation, Nicholson Short-Term Exchange, The Robertson Therapeutic Development Fund, The Denise and Michael Kellen Foundation through the Kellen Women in Science Entrepreneurship Fund, The Alexander Mauro Fellowship, The Danica Foundation, NIH, and Wallenberg AI, and the Autonomous Systems and Software Program (WASP) funded by the Knut and Alice Wallenberg Foundation.