New tool allows users to navigate both full-length protein and domain orthologs.
Genes that originate from a shared ancestor, but separated by a speciation event are termed orthologs. Orthologs provide crucial insights when analysing evolution and inferring protein function. As a result, numerous databases containing ortholog assignments have been developed and applied to multiple areas of life science research. The InParanoid database (InParanoiDB) is an example of an ortholog database. It was developed by the Sonnhammer group at the Department of Biochemistry and Biophysics at Stockholm University (SciLifeLab). It has proven to be especially popular due to the powerful, high-performing ortholog inference capbilites of the underlying algorithm.
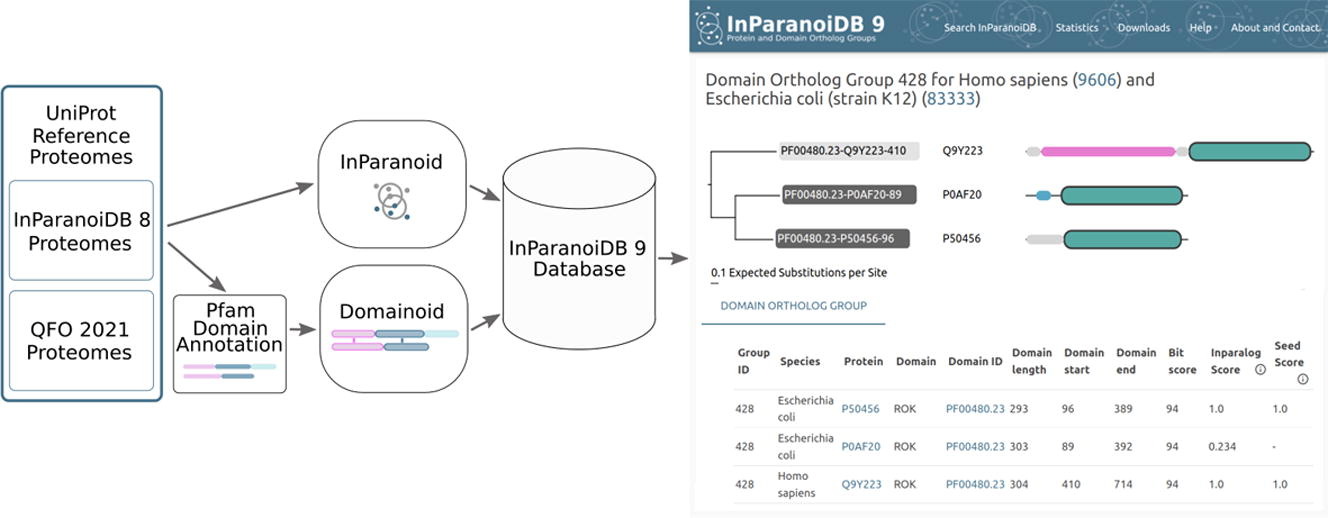
As proteomes become continuously more well-characterised, the expansion and improvement of ortholog databases is both inevitable and essential. The Sonnhammer group has therefore released version 9 of InParanoiDB. This release is based on an improved algorithm to address previous limitations related to predicting orthologous domains, and data on considerably more species and ortholog groups.
The InParanoid-DIAMOND tool was used to build this release. It represents an enhanced version of the algorithm underlying InParanoiDB. It enables orthologs to be inferred more quickly, without loss of accuracy relative to the pre-existing algorithm.
Whilst previous versions of InParanoid comprehensively served the community, there were limitations in predicting othologs at protein domain level. To address the need for domain-level orthology, a new tool, named Domainoid, was developed. The Domainoid tool provides a detailed view on domain level by analysing protein domains defined by Pfam (a protein families database). A comprehensive tool combining the capbilities of both full-length and domain orthologs would serve researchers to carry out their analyses more cohesively.
Version 9 of InParanoiDB includes data from both InParanoid-DIAMOND and Domainoid. It represents an extensive online resource, featuring over 346 million full-length protein ortholog groups and nearly 758 million orthologous domain groups across 640 species. This marks a significant increase in coverage compared to the previous version, which included 273 species and 8 million ortholog groups. The database contains approximately 743 million full-length protein ortholog pairs and over 1.6 billion domain pairs, with more than 6.6 million proteins having at least one ortholog.
The latest version of InParanoiDB allows users to navigate both full-length protein and domain orthologs with an optimised computational pipeline for an efficient and reliable ortholog interface. The associated website supports detailed searches for ortholog groups and displays connections between full-length proteins and their domain counterparts. This includes cases of discordant domain orthology where different domains on the same protein have distinct evolutionary origins. Overall, InParanoiDB 9 offers enhanced coverage and detail, making it a valuable tool for researchers in the field of molecular evolution.
InParanoiDB 9 was built in Python34 (version 3.6.9) using the Django35 (version 3.2.6) framework. All data is stored in a PostgreSQL database. Materialize Css frontend framework was used as a base for the frontend components, and treelib.js was used as a base for drawing trees for each ortholog group. To build multiple sequence alignments and neighbour-joining trees displayed for each ortholog group, Kalign36(version 2.04) and Belvu37 (version 4.44.1b) were used. The InParanoiDB 9 application is run and deployed in a Docker container, managed with Docker Compose (version 3.3).
Article
DOI: 10.1016/j.jmb.2023.168001
Persson, E., & Sonnhammer, E. L. (2023). InParanoiDB 9: ortholog groups for protein domains and full-length proteins. Journal of Molecular Biology, 435(14), 168001.
Funders
This work was supported by the Swedish Research Council Project 2015-05342. Open access funding provided by Stockholm University.
Data Availability
The updated versions of InParanoid-DIAMOND and Domainoid used to obtain the data for InParanoiDB 9 can be found in their respective bitbucket repositories.
Infrastructure
The Department of Biochemistry and Biophysics, Stockholm University and Science for Life Laboratory (Scilifelab) were used for parts of this study.