New data highlight exploring a new metagenomic profiling workflow for ancient DNA
April 15, 2024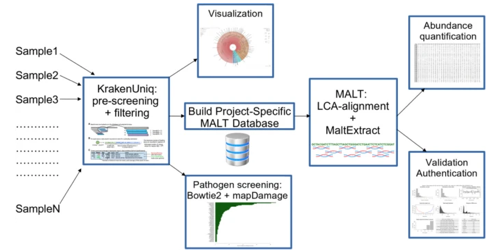
A new data highlight entitled ‘aMeta: a new metagenomic profiling workflow for ancient DNA’ explores a new metagenomic profiling workflow for ancient DNA. It is based on work recently published in Genome Biology by Pochon et al. (2023).
The development of technologies that can be used to study ancient DNA (aDNA) have led to many recent discoveries that allow us a clearer picture of the ancient past. Studies of microbial DNA from archeological samples, and sedimentary ancient DNA (sedaDNA) represent rapidly growing areas of research. However, there remain some limitations in what is possible with aDNA studies. For example, when studying ancient microbial DNA, there are typically high rates of false-positives and false-negatives for the presence of ancient microbial DNA. More stringent ancient metagenomic computational workflows could help to reduce the error rate, but few are available. AMeta represents a new workflow that has been shown to be superior to the current standard.
See the data highlight for more details: aMeta: a new metagenomic profiling workflow for ancient DNA. Relevant data highlights will be shown at the bottom of the page. To get your work featured as a data highlight, please get in touch with us to discuss it.