Background
Proteins are involved in a myriad of biological processes and can perform multiple types of molecular functions. The molecular functions of any given protein are determined by a combination of their inherent properties, environment, and interactions. Since the subcellular location of proteins dictates their environment and possible interactions, it is a key determinant of their molecular functions.
The cellular organisation of proteins has, until recently, been largely unexplored on the global level. The work performed within the SubCellBarCode project (Orre et al., 2019) contributes knowledge of the cellular organisation by providing both a resource for protein subcellular localisation information, and an analysis pipeline/workflow for further large-scale exploration in additional cell types and conditions.
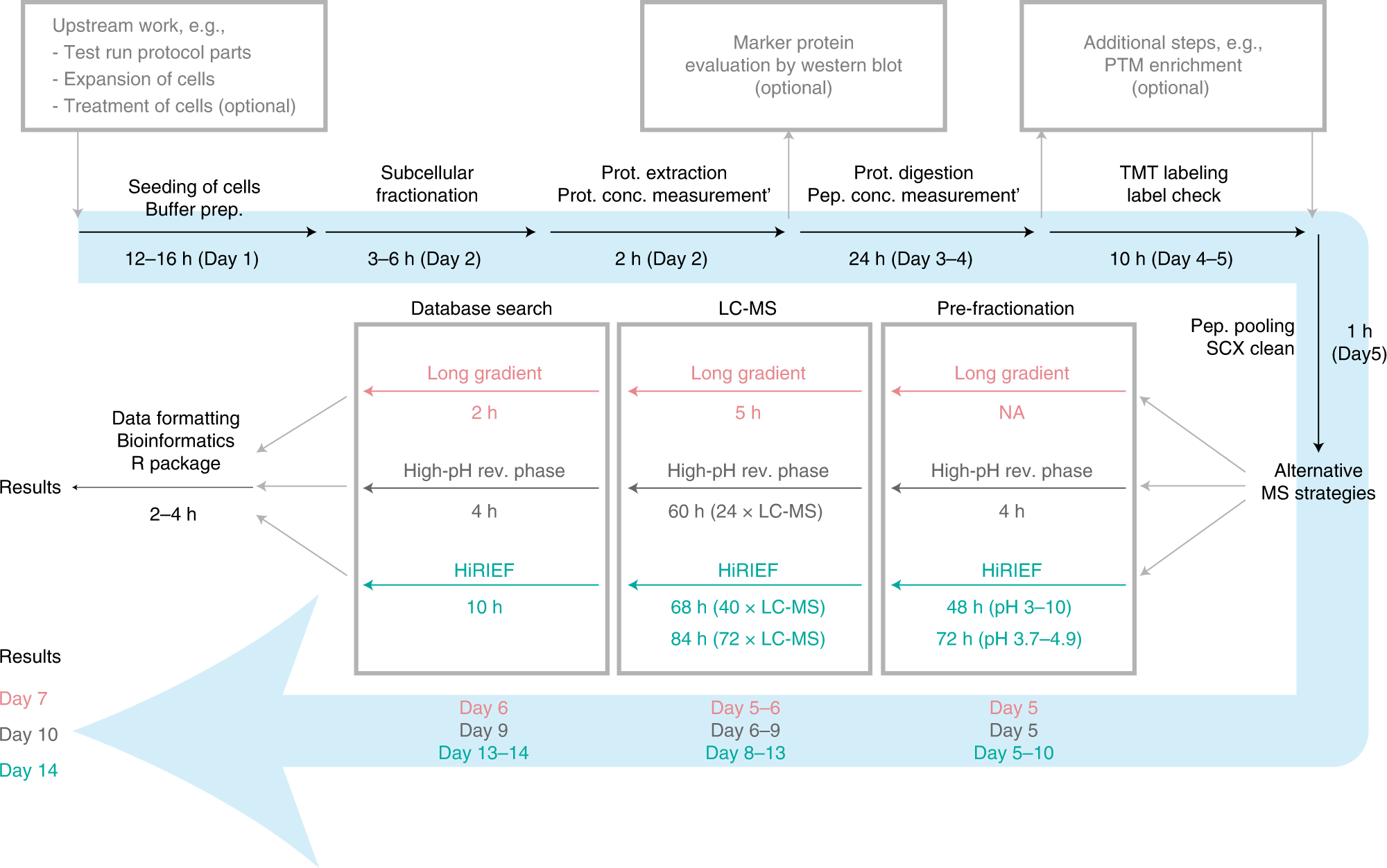
Orre et al. (2019) used cell fractionation together with quantitative mass spectrometry (MS) to study protein subcellular localisation. They then classified protein localisation on a global scale, providing a resource covering proteins mapping to almost 12.5k unique genes across 5 cell lines. They have recently progressed their work by developing an integrated workflow for spatial proteomics research (Arslan & Pan et al., 2022). This comprehensive workflow includes both the wet- and dry-lab steps. The wet-lab protocol encompasses the process from subcellular fractionation to MS sample preparation and analysis. The dry-lab protocol describes all subsequent computational steps required to make determinations about the subcellular locations of proteins. It includes quantitative MS-data analysis, classification based on machine learning, differential localisation analysis, and data visualisation. An overview of the method and approximate timings is shown below.

This section of the SciLifeLab Data Platform has been developed in collaboration with the team working on the SubCellBarCode project. All of the resources produced by the project will be collated here, with the aim of facilitating work on protein subcellular localisation. The page will be continuously updated with the latest outputs from SubCellBarCode and will provide information on how to get support.
SubCellBarCode web portal
The SubCellBarCode web portal includes multiple open-access resources to support spatial proteomics research.
Tools/Apps
There are 4 tools/apps on the web portal:
-
BarCode (single protein localisation) - Explore the subcellular location(s) of 12,418 genes in up to 5 cell lines.
-
CoLocal (correlation network) - Use fractionation profile correlation and classification to evaluate protein co-localisation.
-
NetWork (multiprotein localisation) - Generate localisation networks for proteins with single classified localisation.
-
HeatMap (fractionation profiles) - Explore co-fractionation by clustering protein fractionation profiles.
Each of the tools can be accessed at the bottom of the ‘Home’ page or under the ‘Tools’ subheader. The apps will open at the bottom of the page.
Data
Processed data from the SubCellBarCode project can be queried and downloaded from the SubCellBarCode web portal. It can be accessed by clicking ‘Data’ on the top right hand side of the homepage. Multiple different data types (e.g. fractionation, neighbourhood/compartment predictions, and splice variant localisation) from 5 different cell lines (A431, MCF7, H322, HCC827, and U251) are available. Use the ‘Data Type’ menu to explore a given data type and use the ‘Table’ menu to view data from a given cell line.
Publications
Orre, L. M., Vesterlund, M., Pan, Y., Arslan, T., Zhu, Y., Woodbridge, A. F., Frings, O., Fredlund, E., Lehtiö, J. (2019). SubCellBarCode: proteome-wide mapping of protein localization and relocalization. Molecular Cell 73, P166-182.E7. https://doi.org/10.1016/j.molcel.2018.11.035.
Arslan, T., Pan, Y., Mermelekas, G., Vesterlund, M., Orre, L. M., Lehtiö, J. (2022). SubCellBarCode: integrated workflow for robust spatial proteomics by mass spectrometry. Nature Protocols 17, 1832-1867. https://doi.org/10.1038/s41596-022-00699-2.
Available code
The SubCellBarCode team have created an R-package that can be used to perform the dry-lab portion of their protocol, as described in Arslan & Pan et al. (2022). The R-package is freely available at Bioconductor.
The SubCellBarCode pipeline used in Arslan & Pan et al. (2022) can also be found on GitHub.
Available data
The mass spectrometry (MS) proteomics data for the analysis of the HeLa cell line are available in the ProtomeXchange Consortium via the jPOST partner repository (identifier: PXD022533).
The MS proteomics data for the analysis of the original 5 cell lines (A431, MCF7, H322, HCC827, and U251) was deposited to the ProteomeXchange Consortium via the PRIDE partner repository (identifier: PXD006895).
Processed data from the SubCellBarCode project can be queried and downloaded from the SubCellBarCode web portal. See the web portal data subsection above for details.
Additional resources
Contact for support
A contact form is available on the SubCellBarCode web portal. Click on the email symbol on the bottom right of the homepage to access it.
Alternatively, it is possible to directly email Lukas Orre or Janne Lehtiö, both researchers involved in the project, with any questions.