‘SubCellBarCode’ – a subcellular proteome resource and analysis pipeline now available on the Data Platform
The subcellular location of a protein is a major determining factor of that protein’s function. Understanding the subcellular distribution of proteins within cells is thus crucial for elucidating the biology of that cell. Unsurprisingly, the subcellular location of proteins is of great importance to multiple fields within biology and medicine. For example, aberrant localisation of proteins has been linked to cancer and neurodegenerative diseases. Consequently, determination of protein localisation has been the subject of multiple studies and large-scale efforts. However, still further knowledge is needed in order to get a global view of cellular proteome organisation.
To date, two main types of methods have been used to study protein localisation; fractionation-based methods and imaging-based methods. Imaging-based methods visualise proteins directly in their environment (in situ) and can even be used to perform in vivo studies. They can, however, be biased by cross-reactivity and the presence of artefacts that, in turn, may influence data integrity. In addition, imaging-based methods are typically time- and labour-intensive, as well as expensive to use. By contrast, fractionation-based methods involve experimental separation of cellular compartments based on physical properties and subsequent quantification of proteins in the generated fractions. Recent technological advances in instrumentation and machine learning have made fractionation-based methods increasingly more effective.
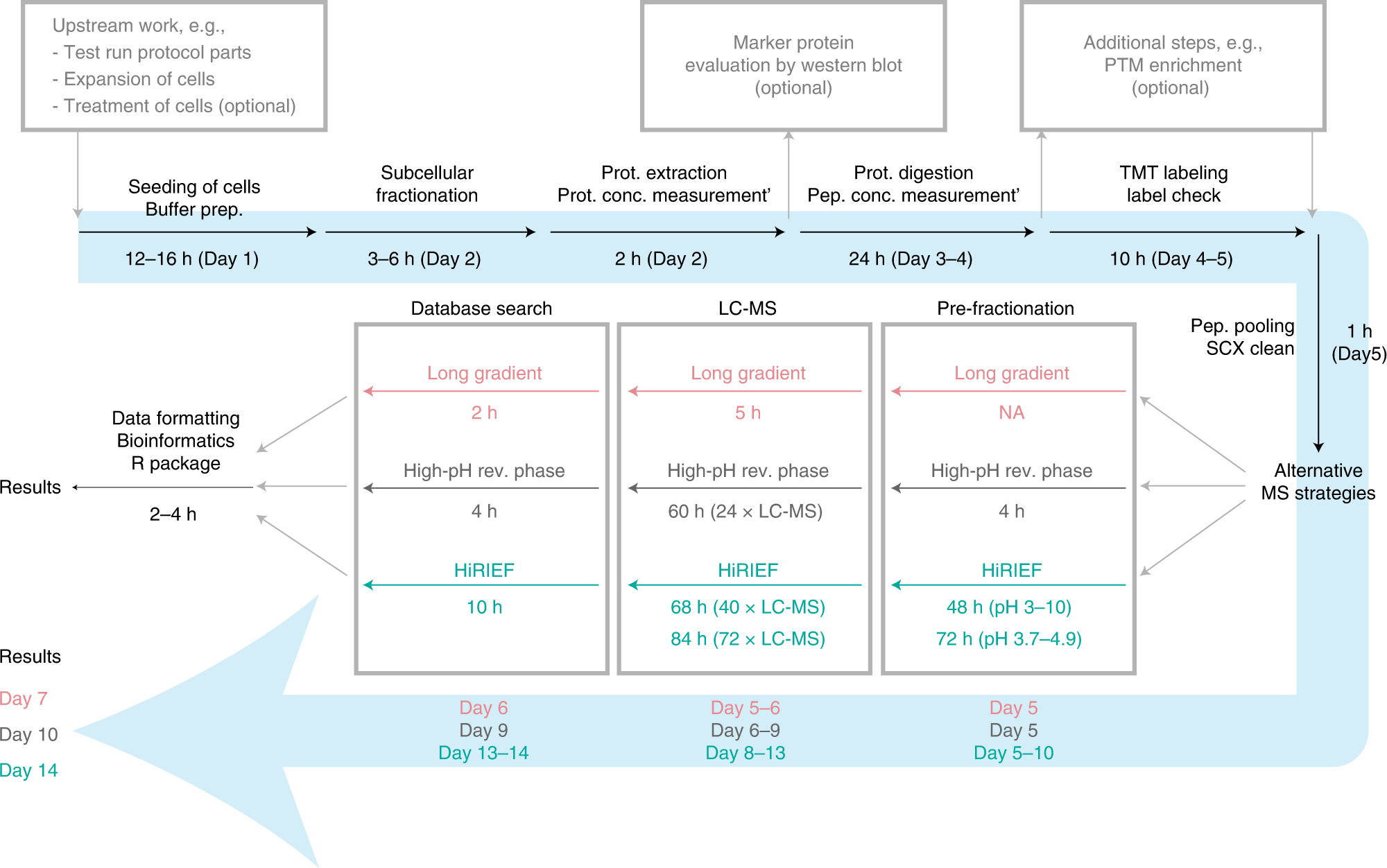
Orre and colleagues at Karolinska Institutet/SciLifeLab have developed a robust pipeline for fractionation-based subcellular proteome analysis, termed ‘SubCellBarCode’ (Orre et al., 2019; Arslan & Pan et al., 2022). The SubCellBarCode pipeline comprises three parts. In the first part, cells are divided into five discrete cellular fractions by extraction of cytosolic proteins and sequential centrifugation-based isolation of the different cellular compartments. The next part involves protein extraction and mass spectrometry (MS)-based relative quantification of proteins across the different fractions. The third and final part is centred around bioinformatic analysis and the classification of protein subcellular localisation using the SubCellBarCode R package. Stacked bar plots are used to visualise the resulting probabilities of each of the protein’s subcellular localisation. These plots resemble barcodes, uniquely representing the proteins, and are what gave the method its name.
In their first study, Orre and colleagues (2019) used the SubCellBarCode pipeline to obtain a proteome-wide view of protein subcellular location. In total, the localisation of more than 12,000 proteins were mapped across five human cancer cell lines (epidermoid carcinoma A431, glioblastoma U251, breast cancer MCF7, and lung cancer NCI-H322 and HCC-827). The pipeline accurately classifies protein subcellular location, as shown by a high level of consistency between classifications made using this pipeline and the available public domain data. The researchers also used the pipeline to study a number of research questions related to the subcellular localisation of proteins. This provided important new insights into both the cellular architecture and spatial organisation of the proteome. For example, the results showed that most proteins have a single primary subcellular location and that subcellular location was rarely affected by alternative splicing. Importantly, the researchers also showed that the method could be used for proteome-wide analysis of protein relocalisation in response to drug treatment. This indicates that the method can contribute knowledge in studies on drug mechanism of action and cell response. The results and analysis tools generated by the SubCellBarCode project were made available by the research team on a custom web platform.
The same research group have recently published a Nature Protocols paper describing the details of the SubCellBarCode pipeline (Arslan & Pan et al., 2022). This paper provides a comprehensive overview of all of the steps of the pipeline, from the wet-lab methodology to completing the dry-lab computational analysis. The wet-lab portion details how to complete subcellular fractionation, and MS sample preparation and analysis. It contains descriptions of the materials, equipment, software, and general set-up needed for the pipeline, as well as providing information about the level of training and time required to carry out the steps. In accordance with open science, the researchers have openly shared data and code in GitHub, Bioconductor, and PRIDE. In the dry-lab portion of the pipeline, the researchers explain how to complete subsequent quantitative MS-data analysis, machine learning-based classification, differential localisation analysis, and data visualisation steps. The detailed protocol provided by the researchers in this latest paper facilitates the reuse of this pipeline and will thus hopefully contribute to future research on protein subcellular location.
In summary, the team behind the SubCellBarCode project have shown that their pipeline is simple, yet robust. A major advantage compared to previous methods used in this area is that all steps, from starting with cell harvest to finished classification output, can be performed within 1-2 weeks.
All of the data, code, protocols and additional resources produced by the SubCellBarCode project have now been aggregated on the SciLifeLab Data Platform as a service page. Please see the SubCellBarCode service page for more. It is hoped that this will further encourage a more widespread adoption of this pipeline throughout broader scientific community. The dedicated area on the SciLifeLab Data Platform will not be static, as it will be continuously updated with new developments from the project.
Data and Code
Information about all of the data, code, and other resources related to the SubCellBarCode project can be found in the SubCellBarCode service page.
Articles
Orre, L. M., Vesterlund, M., Pan, Y., Arslan, T., Zhu, Y., Woodbridge, A. F., Frings, O., Fredlund, E., Lehtiö, J. (2019). SubCellBarCode: proteome-wide mapping of protein localization and relocalization. Molecular Cell 73, P166-182.E7. https://doi.org/10.1016/j.molcel.2018.11.035.
Arslan, T., Pan, Y., Mermelekas, G., Vesterlund, M., Orre, L. M., Lehtiö, J. (2022). SubCellBarCode: integrated workflow for robust spatial proteomics by mass spectrometry. Nature Protocols 17, 1832-1867. https://doi.org/10.1038/s41596-022-00699-2.
Funding
The research was supported by the Swedish Foundation for Strategic Research, Swedish Cancer Society, Swedish Research Council, Swedish Childhood Cancer Foundation, The Cancer Research Funds of Radiumhemmet and Stockholm’s County Council.