New tool explores multi-omic analyses of childhood ALL cancer to identify putative drug candidates
Precision medicine involves the use of genetic information from the individual’s own genes and proteins to enable disease diagnosis, treatment, and prevention. It is especially important in treatment of cancer. Precision medicine enables, for example, the use of therapies that specifically target cancer cells, and the use of tumor biomarkers to diagnose specific cancers. It can also involve the development of novel diagnostic tools for research on cancer, as well as other diseases.
Each year about 300 children in Sweden are diagnosed with cancer. Acute lymphoblastic leukemia (ALL) is the most common form of childhood cancer, accounting for one in three childhood cancer diagnoses. Survival rates for ALL have improved in recent years due to the development of a number of different chemotherapy protocols and allogeneic hematopoietic stem cell transplantation (HSCT). Nevertheless, between 15 and 20% of children either do not respond well to currently available treatments or relapse, typically with poor survival outcomes. In addition, 80% of childhood ALL survivors experience a serious post-treatment medical event by middle age. In recent years, targeted therapies have been developed that reduce post-treatment complications and improve the outcome. However, work is still very much needed to develop novel therapeutic modalities for the treatment of high-risk patients and patients who relapse.
The development of novel pharmacological and cellular therapies will require further research into the biological differences between children that are responsive to treatment, and those are not. Previous research using genetic and transcriptomic profiling have been able to, in part, explain why some children respond well and some do not. However, it is hoped that precision medicine will give better insight, as it will enable the characterization of the biomarkers and biology of responsive and non-responsive subtypes. This will be especially important for gaining more information on non-responders, about which relatively less is understood.
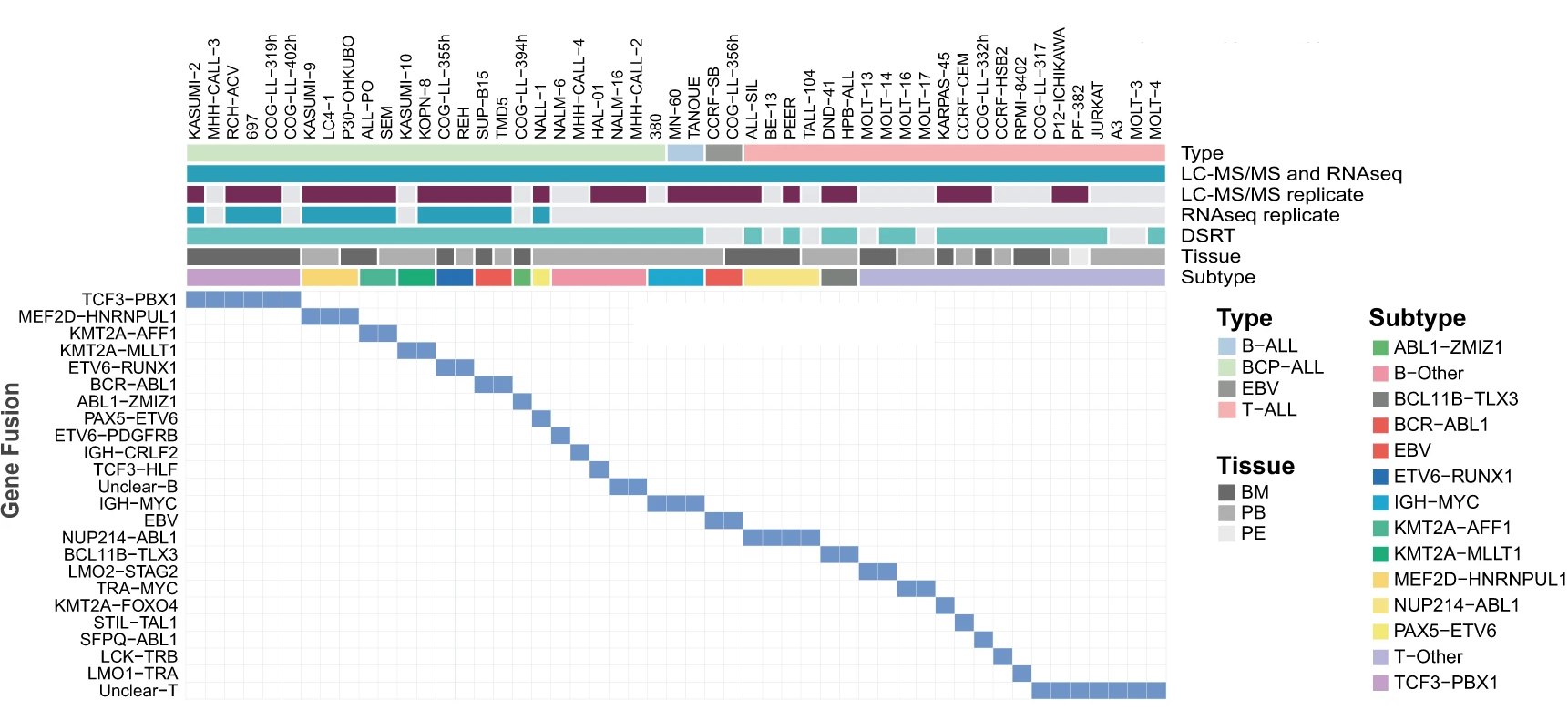
In a recently published article, researchers from SciLifeLab, Karolinska Institute and University of Muenster (first authors: Isabelle Rose Leo, Luay Aswad, Matthias Stahl; corresponding author: Rozbeh Jafari) have investigated methods to identify potential drug candidates for childhood ALL. The researchers adopted a comprehensive multi-omic analysis of 49 readily available childhood ALL cell lines, using proteomics, transcriptomics, and pharmacoproteomic characterization.
Leo, Aswad, Stahl and colleagues used multi-omics layered analysis, including RNAseq, liquid chromatography-mass spectrometry (LC-MS), and drug sensitivity and resistance testing (DSRT), to study readily available childhood ALL cell lines. First, the research collaboration quantified over 12,000 proteins and 19,000 protein-coding transcripts. The sensitivity of 43 ALL cell lines to a panel of 528 approved oncological drugs and potential drug candidates was then tested using the DSRT.
The dataset could be used to identify changes in how efficiently proteins are produced and how stable they are in cells, based on comparing differences in abundance of RNA transcripts versus proteins. The authors also analyzed protein abundances to determine drug mechanisms, and identified that some unexpected proteins were linked to drug sensitivity, which indicates secondary drug targets that could be important tools in future treatments. Additionally, by comparing many cell lines with different lineages and genetic fusions, the data could be used to identify biological processes and drug vulnerabilities that are specifically important within subtypes of ALL.
The researchers used the approach outlined above to identify a putative therapeutic candidate for a high-risk subtype of ALL. Children with ALL that possess the MEF2D-HNRNPUL1 fusion subtype are known to be at greater risk of relapse and a lower chance of survival than children with other types of ALL. The therapeutic candidate for potentially better treatment for such high-risk subtypes is called diacylglycerol-analog bryostatin-1, which is an immunomodulatory drug. It can activate the proapoptotic ERK signaling pathway in early pre-B cells. As the safety of this drug for use in humans has been established using phase I and Phase II drug testing, and pharmacological profile is already known, it could easily be repurposed to carry out animal studies of ALL.
“With this dataset, we have demonstrated that disease-specific in-depth representation is highly valuable and adds additional layers of knowledge. We are continuing to apply these methods to additional translational pediatric ALL models to expand the FORALL resource, and we will also explore drug sensitivity results in depth in future studies” says Rozbeh Jafari, the senior corresponding author of the study.
In summary, multi-omics approaches can be applied to childhood ALL lines for the purposes of potential drug discovery. Such approaches have relevance to precision medicine and can be used to better understand specific mechanisms of drug sensitivity in childhood ALL.
Leo, Aswad, Stahl and colleagues have made their work available as an user-friendly web application (R Shiny app) called FORALL (Functional Omics Research of Acute Lymphoblastic Leukemia). The web application makes it possible for other researchers to further explore the proteomic, transcriptomic, and drug sensitivity data described in the study.
Data
- FORALL web portal, available at https://proteomics.se/forall:
- Proteomics: MS-based protein profiles of 49 childhood ALL cell lines and two EBV transformed B-cell lines with biological replicates (N=82 samples).
- Transcriptomics: RNA-seq-based total RNA profiles of 49 childhood ALL cell lines and two as two EBV transformed cell lines with biological replicates (N=66 samples).
- Drugs response: 25 BCP-ALL, 16 T-ALL, and 2 B-ALL cell lines responses to treatment of a panel of 528 drugs.
- Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD023662.
- Raw RNA-seq data generated in this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE168386.
- Publicly available transcriptomic data were obtained from The European Genome-phenome Archive (EGA) (Dataset ID: EGAD00001002704 and EGAD00001002692) after access was provided from St. Jude Children’s Research Hospital Memphis - Washington University Pediatric Cancer Genome Project.
- Raw viable cell count output from flow cytometry experiments can be found on Github.
Article
Leo, I. R., Aswad, L., Stahl, M., Kunold, E., Post, F., Erkers, T., Struyf, N., Mermelekas, G., Joshi, R. N., Gracia-Villacampa, E., Östling, P., Kallioniemi, O. P., Tamm, K. P., Siavelis, I., Lehtiö, J., Vesterlund, M.Jafari, R. (2022). Integrative multi-omics and drug response profiling of childhood acute lymphoblastic leukemia cell lines. Nature Commununications, 13, 1691. 10.1038/s41467-022-29224-5.
Funding
This study was supported by grants from the Swedish Childhood Cancer Foundation, the Swedish Research Council, Felix Mindus Contribution to Leukemia research, Dr. Åke Olsson Foundation for Hematological Research, Cancer Society Stockholm and the King Gustaf V Jubilee Fund, and Magnus Bergvalls Stiftelse.