Here we showcase exceptional, recently published open source tools and open data that could be useful for data-driven life science research, as well as research resulting from their use. The Data Highlights are written by the SciLifeLab Data Platform editorial team or external contributors.
October 16, 2025
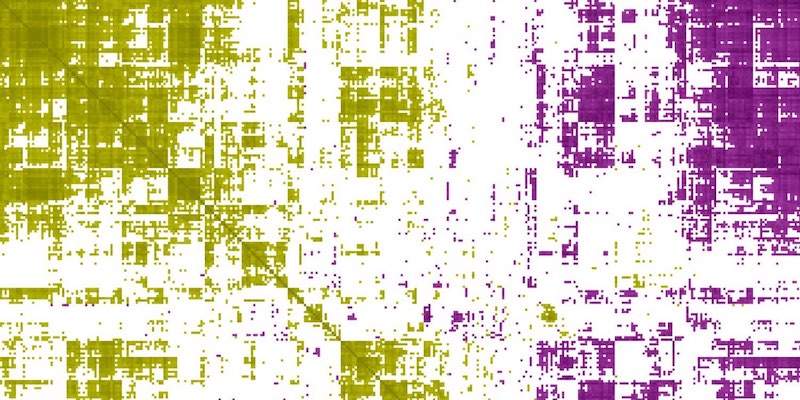
FunCoup 6 bridges the gap between functional association and regulatory networks across many organisms, enabling users to explore regulation and module-level insights in a unified framework.
September 18, 2025
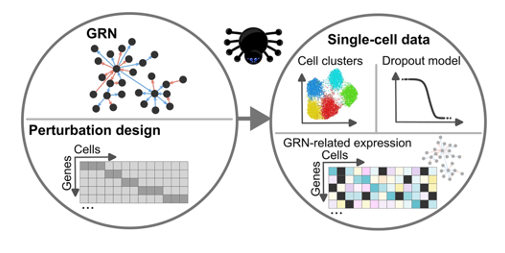
GeneSPIDER2 allows unique capacity to simulate perturbation-aware single-cell datasets. This aligns well with the objectives of data platforms supporting systems medicine, pathogen research, and pandemic preparedness.
November 7, 2024
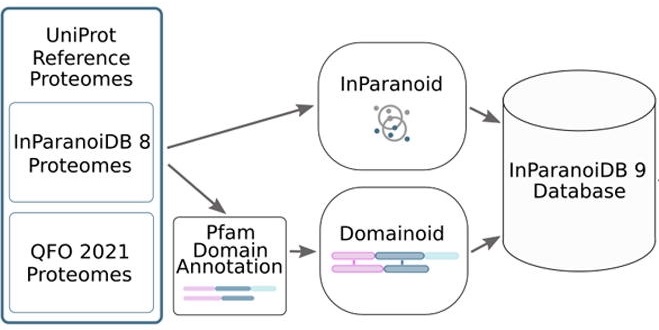
The latest version of InParanoiDB by the Sonnhammer group allows users to navigate both full-length protein and domain orthologs with an optimised computational pipeline for an efficient and reliable ortholog interface.
August 19, 2024
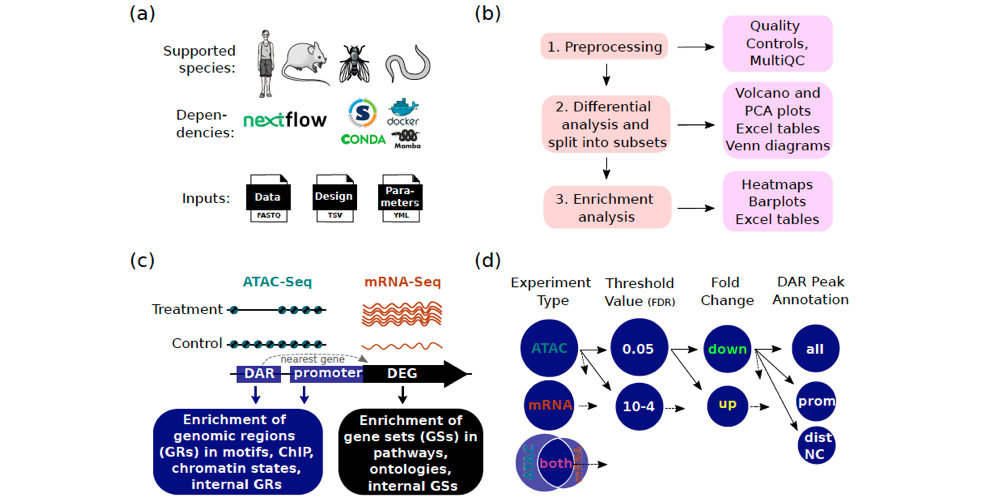
Salignon et al. created Cactus, a new pipeline that can be used for comprehensive ATAC-Seq and mRNA-Seq data analysis. Cactus contains multiple unique functions compared to other, similar pipelines, e.g. enrichment in chromatin states and ChIP-Seq binding sites.
April 15, 2024
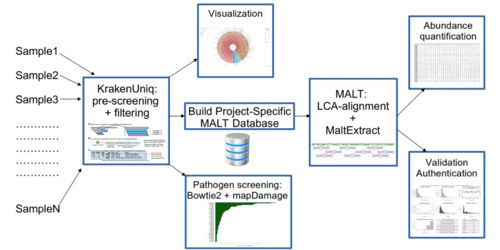
Pochon et al. have developed aMeta; a new metagenomic profiling workflow for ancient DNA. aMeta was found to have superior microbial detection, and require less computer memory than the workflow currently considered the de facto standard.
January 19, 2024
Babačić and colleagues expanded the coverage of the soluble blood proteome using mass spectrometry. In order to support further research in this area, their results have been added to an open-access app.
January 19, 2024
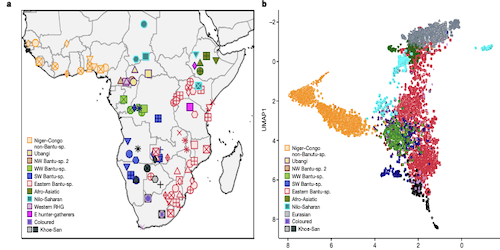
This study by Cesar A. Fortes-Lima, Concetta Burgarella, Rickard Hammarén et al is a comprehensive investigation of the genetic legacy of the Bantu expansion of the genomes of Bantu speaking populations today. The authors make avilable both genotyping information for 1,763 African individuals and whole genomes for 12 Late Iron Age individuals.
December 20, 2023
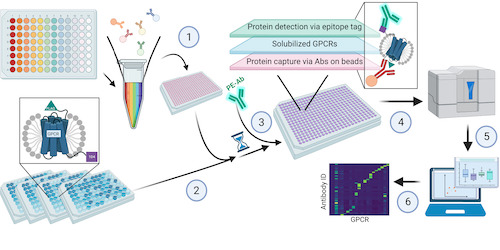
This study by Dahl, Kotilar and Bendes et al address this challenge of developing a high-throughput method to study GPCRs. Data and app shared.
September 19, 2023
This study by Knöppel, Broström et al is a large effort to elucidate replication initiation in bacteria. The authors have openly shared over 3 TB of microscopy imaging data.
June 2, 2023
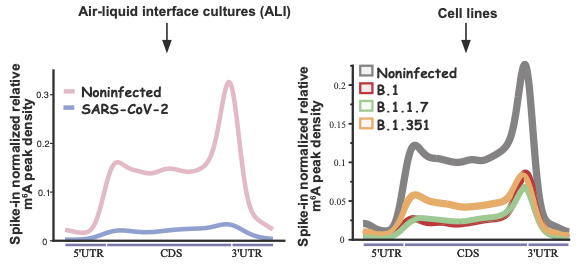
Vaid and collaborators studied how the gene expression profile of m6A mRNA is affected both during and after COVID-19 infection. All sequencing data and the source code for analysis are shared.
May 5, 2023
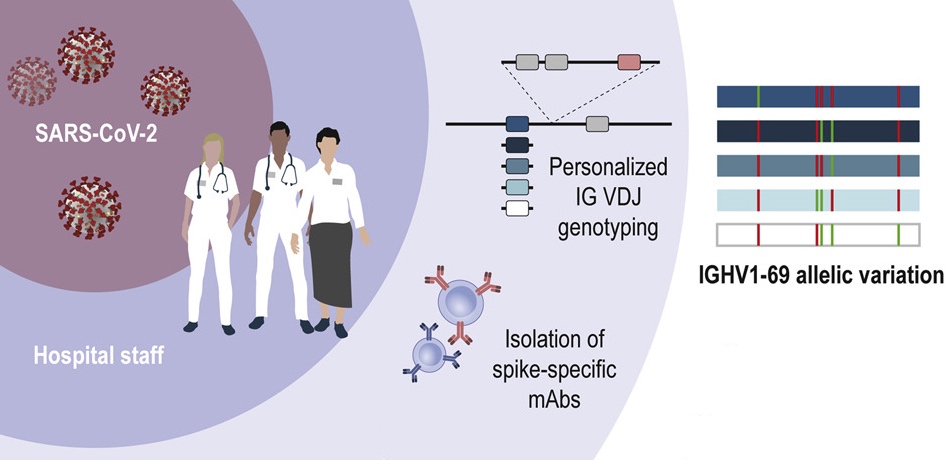
Pushparaj and colleagues use genotyping and haplotype analysis to show high genetic diversity in IGH genes among humans, which may influence our response to infections. Data, and IgDiscover software shared.
April 18, 2023
Recent study from Elf lab at Uppsala University/SciLifeLab shows perseverance can be a reason for antibiotic resistance development in.E coli. Image data shared in SciLifeLab Data Repository.
February 24, 2023
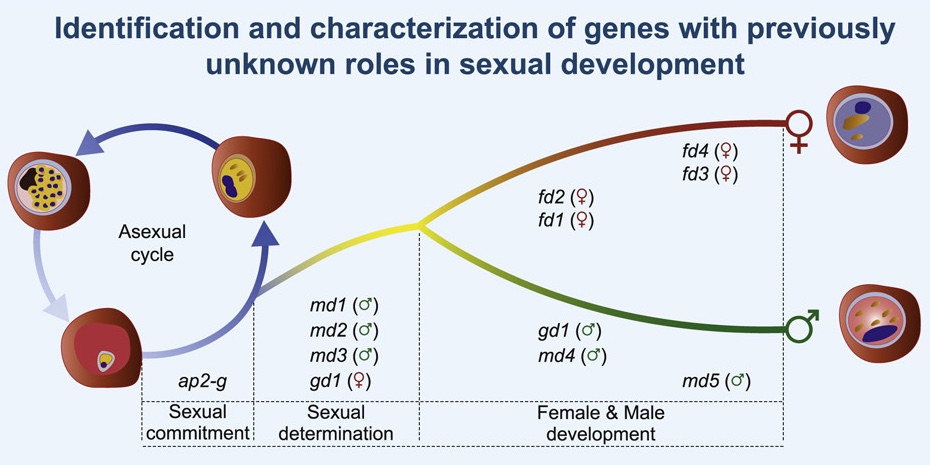
Russell et al. (2023) details the discovery of sex determination genes in the malaria parasite Plasmodium berghei, and how they are essential in transmission of malaria via mosquitoes. Data and code shared openly.
January 20, 2023
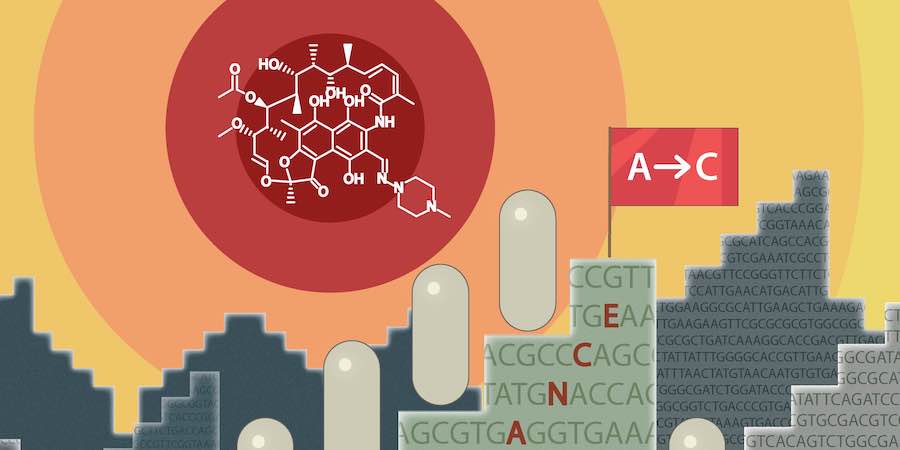
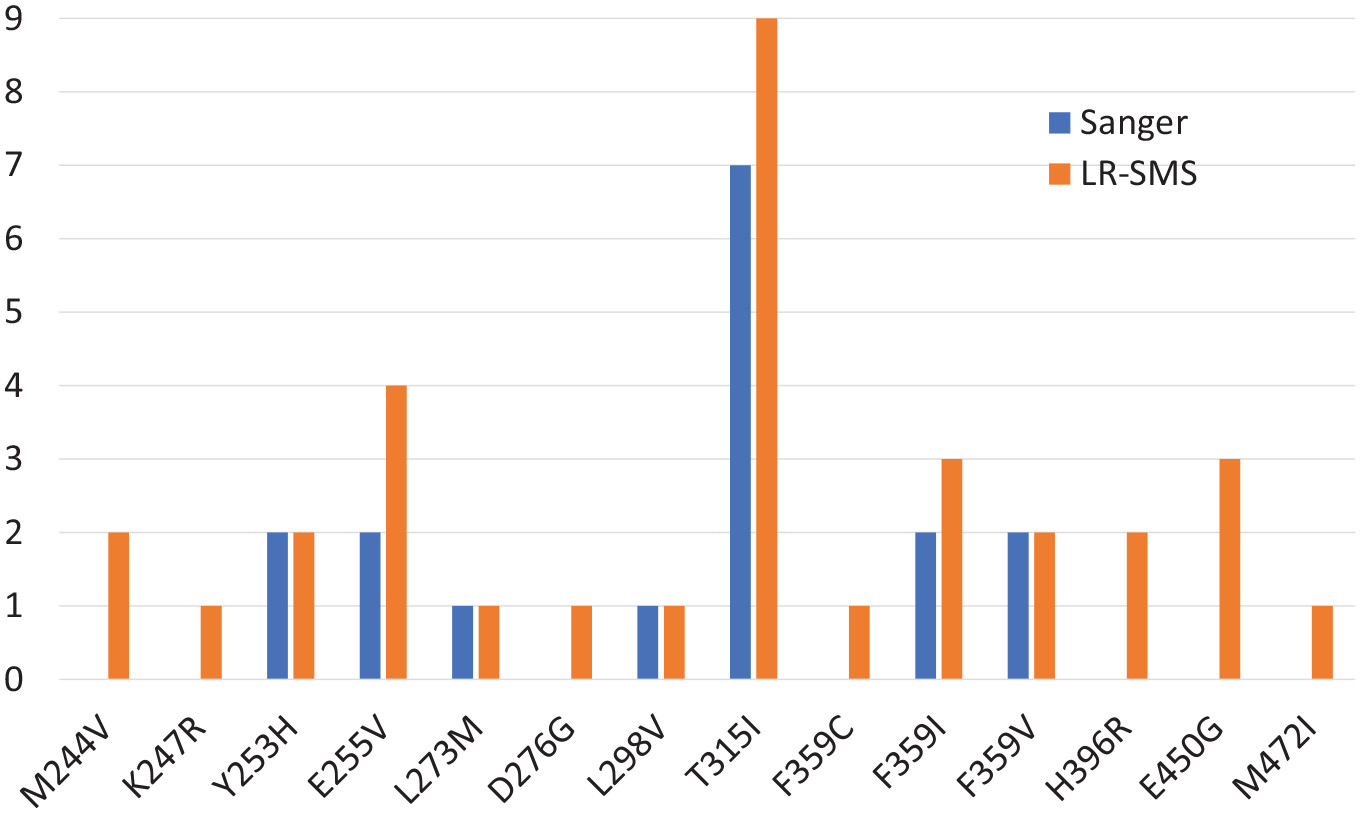
Schaal et al. (2022) found that long-read sequencing was more sensitive than Sanger sequencing for detecting mutations associated with the development of resistance to certain cancer drugs. Custom software produced as part of this study is shared openly.
December 19, 2022
Dahmane et al (2022) used cryo-electron tomography to provide an integrated structural framework for multiple stages of the poliovirus life cycle. Data and code are shared openly.
October 21, 2022
Study by Jamy et al. show that hundreds of transitions have occurred between marine and non-marine habitats over the course of two billion years of eukaryotic evolution. Code and data are shared openly.
October 13, 2022
The GenErode pipeline is the first bioinformatics pipeline that can process and analyse ancient, historical, and modern sequencing data from the same species with the aim of generating comparable estimates of genomic erosion indices.
October 6, 2022
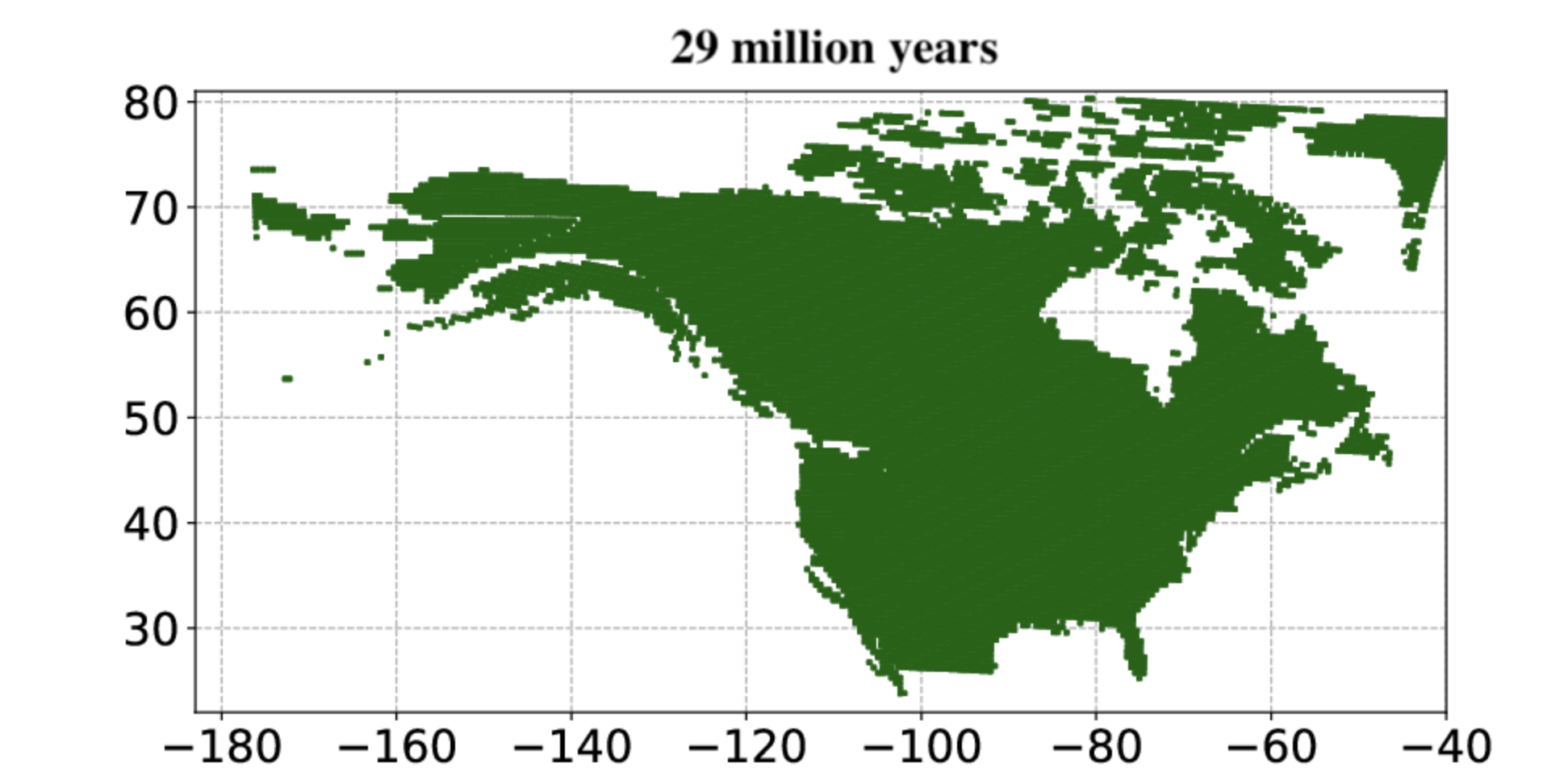
The study uses a Bayesian deep-learning model trained using fossil evidence of mammal-plant interactions to explore the origin and expansion of open grassland in North America. The study used open data sources, and openly shares the code and functionalities produced.
September 29, 2022
The SubCellBarCode project focusses on determining the subcellular localisation of proteins. We explore two major publications produced by the project in detail as well as the resources openly shared from it.
August 12, 2022
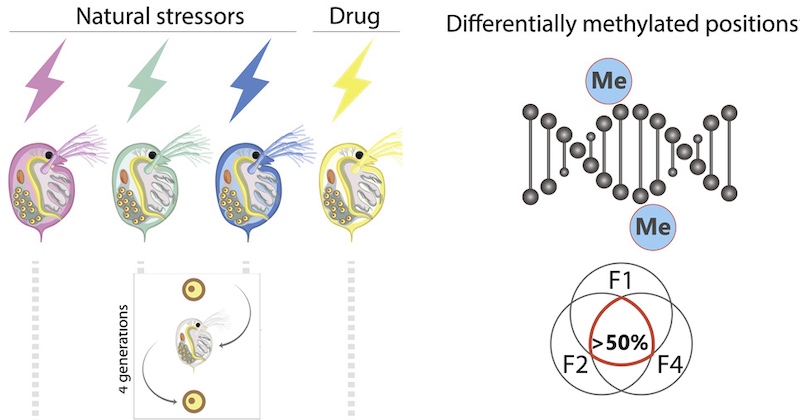
New study from Feiner and colleagues shows that environmental stressors induce DNA methylation in D. magna and that this epigenetic effect is heritable. Sequence data and code to analyse DNA-methylation and fitness are openly shared.
July 7, 2022
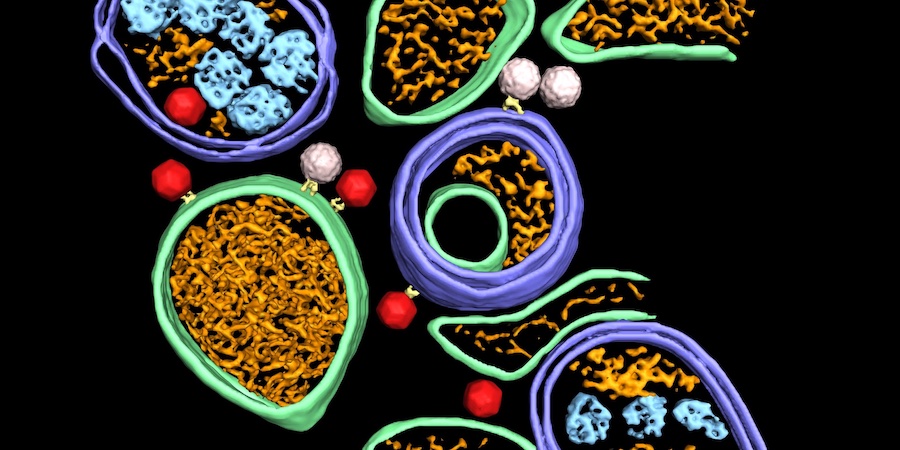
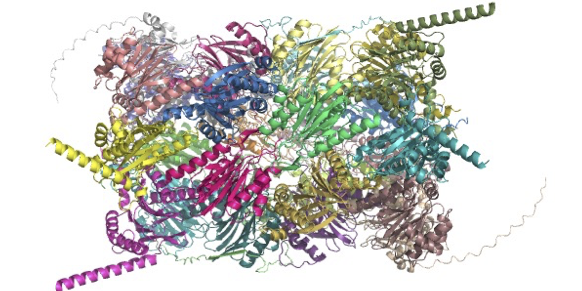
Recent publication shows how scientists at SciLifeLab use cryo-EM to reveal the sequential steps involved in mitoribosome assembly. Data has been made publicly available.
June 21, 2022
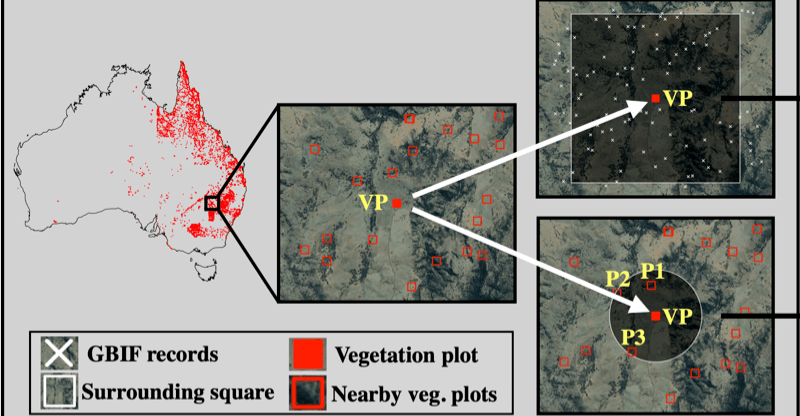
The study shows that deep learning frameworks can be used as a viable approach to estimate patterns of biodiversity over a large area. Publicly available data sources were used in the study, all code related to the deep learning framework is openly shared.
May 10, 2022
Leo and colleagues used multi-omic analysis for studies of childhood Acute lymphoblastic leukemia (ALL) . New tool for improving childhood ALL cancer treatment developed and shared.
May 6, 2022
Study gives insight into host-viral interactions of Crimean-Congo hemorrhagic fever, an infectious disease without available treatments. Raw RNAseq, mass spectrometry proteomics data, and code shared.
May 5, 2022
New pipeline called FoldDock uses AlphaFold2 to provide accurate predictions of heterodimeric complexes structures. This pipeline has the potential for rapidly expanding knowledge about structural protein interactions at a low cost. The code required to run FoldDock and reproduce the analysis has been published on GitLab.
April 25, 2022
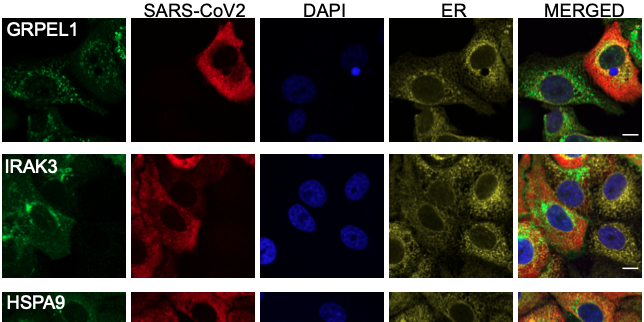
Expression of 602 host proteins were evaluated in populations of infected and non-infected cells using immunofluorescence. ≈75,000 images have been published as a resource for further studies.